1/25/18: NOTE: Since the November release of the CheXNet paper on ArXiV, there has been a healthy and extensive online discussion on twitter, reddit, and online blogs. The Stanford paper has undergone at least two revisions with some substantial modifications, most importantly the replacement of ROC curves with F1 scores and a bootstrap calculation of significance. Some details about the methodology which were not released in the original version have come out, particularly the “re-labeling” of ground truth by Stanford radiologists. My comment about the thoracic specialist has completely borne out on further release of information. And the problems with ChestXRay14’s labeling (why the Stanford docs re-labeled) are now well-known.
The investigation and discussion of this paper has been spearheaded by Luke Oaken Rayner, who has spent months corresponding with the author and discussing the paper. For further Information, see below.
The discussion on CheXNet appears to be over, and there has been a great deal of collective learning in it. The Stanford group should be lauded for their willingness to engage in open peer review and modify their paper substantially after it. There is no question that a typical 18-24 month process of review and discussion was fast-tracked in the last two months. Relevant blog links are below after my December addendum. This will be my last update on this post, as it is “not so brief” any longer!
Andrew Ng released CheXNet yesterday on ArXiv (citation) and promoted it with a tweet which caused a bit of a stir on the internet and related radiology social media sites like Aunt Minnie. Before Radiologists throw away their board certifications and look for jobs as Uber drivers, a few comments on what this does and does not do.
First off, from the Machine Learning perspective, methodologies check out. It uses a 121 layer DenseNet, which is a powerful convolutional neural network. While code has not yet been provided, the DenseNet seems similar to code repositories online where 121 layers are a pre-made format. 80/20 split for Training/Validation seems pretty reasonable (from my friend, Kirk Borne), Random initialization, minibatches of 16 w/oversampling positive classes, and a progressively decaying validation loss are utilized, all of which are pretty standard. Class activation mappings are used to visualize areas in the image most indicative of the activated class (in this case, pneumonia). This is an interesting technique that can be used to provide some human-interpretable insights into the potentially opaque DenseNet.
The last Fully Connected (FC) layer is replaced by a single output (only one class is being tested for – pneumonia) coupled to a sigmoid function (an activation function – see here) to give a probability between 0 and 1. Again, pretty standard for a binary classification. The multiclass portion of the study was performed seperately/later.
The test portion of the study was 420 Chest X-rays read by four radiologists, one of whom was a thoracic specialist. They could choose between the 14 pathologies in the ChestX-ray14 dataset, read blind without any clinical data.
So, a ROC curve was created, showing three radiologists similar to each other, and one outlier.The radiologists lie slightly under the ROC curve of the CheXNet classifier. But, a miss is as good as a mile, so the claims of at or above radiologist performance are accurate, because math. As Luke Oakden Rayner points out, this would probably not pass statistical muster.
So that’s the study. Now, I will pick some bones with the study.
First, only including one thoracic radiologist is relevant, if you are going to make ground truth agreement of 3 out of four radiologists. (Addendum: And, for statistical and methodological reasons discussed online, the 3 out of 4 implementation was initially flawed as scored) General radiologists will be less specific than specialist radiologists, and that is one of the reasons why we have moved to specialty-specific reads over the last 20 years. If the three general rads disagreed with the thoracic rad, the thoracic rad’s ground truth would be discarded. Think about this – you would take the word of the generalist over the specialist, despite greater training. (1/25 Addendum: proven right on this one. The thoracic radiologist is an outlier with a higher F1 score) Even Google didn’t do this in their retinal machine learning paper. Instead, Google used their three retinal specialists as ground truth and then looked at how the non-specialty opthalmologists were able to evaluate that data and what it meant to the training dataset. (Thanks, Melody!) Nevertheless, all rads lie reasonably along the same ROC curve, so methodologically it checks out the radiologists are likely of equal ability but different sensitivities/specificities.
Second, the Wang ChestXray14 dataset is a dataset that was data-mined from NIH radiology reports. This means that for the dataset, ground truth was whatever the radiologists said it was. I’m not casting aspersions on the NIH radiologists, as I am sure they are pretty good. I’m simply saying that the dataset’s ground truth is what it says it is, not necessarily what the patient’s clinical condition was. As proof of that, here are a few cells from the findings field on this dataset.

In any case, the NIH radiologists more than a few times perhaps couldn’t tell either, or identified one finding as the cause of the other (Infiltrate & Pneumonia mentioned side by side) and at the top you have the three fields “atelectasis” “consolidation” & “Pneumonia” – is this concurrent pneumonia with consolidation with some atelectasis elsewhere, or is it “atelectasis vs consolidation cannot r/o pneumonia” (as radiologists we say these things). While the text miner purports to use several advanced NLP tools to avoid these kinds of problems, in practice it does not seem to do so. (See addendum below, further addendum, confirmed by Jeremy Howard) Dr. Ng, if you read this, I have the utmost respect for you and your team, and I have learned from you. But I would love to know your rebuttal, and I would urge you to publish those results. Or perhaps someone should do it for reproducibility purposes.
Finally, I’m bringing up these points not to be a killjoy, but to be balanced. I think it is important to see this and prevent someone from making a really boneheaded decision of firing their radiologists to put in a computer diagnostic system (not in the US, but elsewhere) and realizing it doesn’t work after spending a vast sum of money on it. Startups competing in the field who do not have deep healthcare experience need to be aware of potential pitfalls in their product. I’m saying this because real people could be really hurt and impacted if we don’t manage this transition into AI well. Maybe all parties involved in medical image analysis should join us in taking the Hippocratic Oath, CEO’s and developers included.
Thanks for reading, and feel free to comment here or on twitter or connect on linkedin to me: @drsxr
December Addendum: ChestX-ray14 is based on the ChestX-ray8 database which is described in a paper released on ArXiv by Xiaosong Wang et al. The text mining is based upon a hand-crafted rule-based parser using weak labeling designed to account for “negation & uncertainty”, not merely application of regular expressions. Relationships between multiple labels are expressed, and while labels can stand alone, for the label ‘pneumonia’, the most common associated label is ‘infiltrate’. A graph showing relationships between the different labels in the dataset is here (from Wang Et Al.)
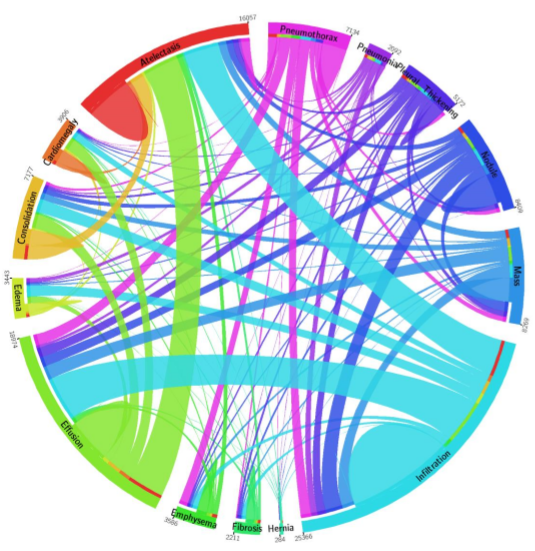
Pneumonia is purple with 2062 cases, and one can see the largest association is with infiltration, then edema and effusion. A few associations with atelectasis also exist (thinner line).
The dataset methodology claims to account for these issues at up to 90% precision reported in ChestX-ray8, with similar precision inferred in ChestX-ray14.
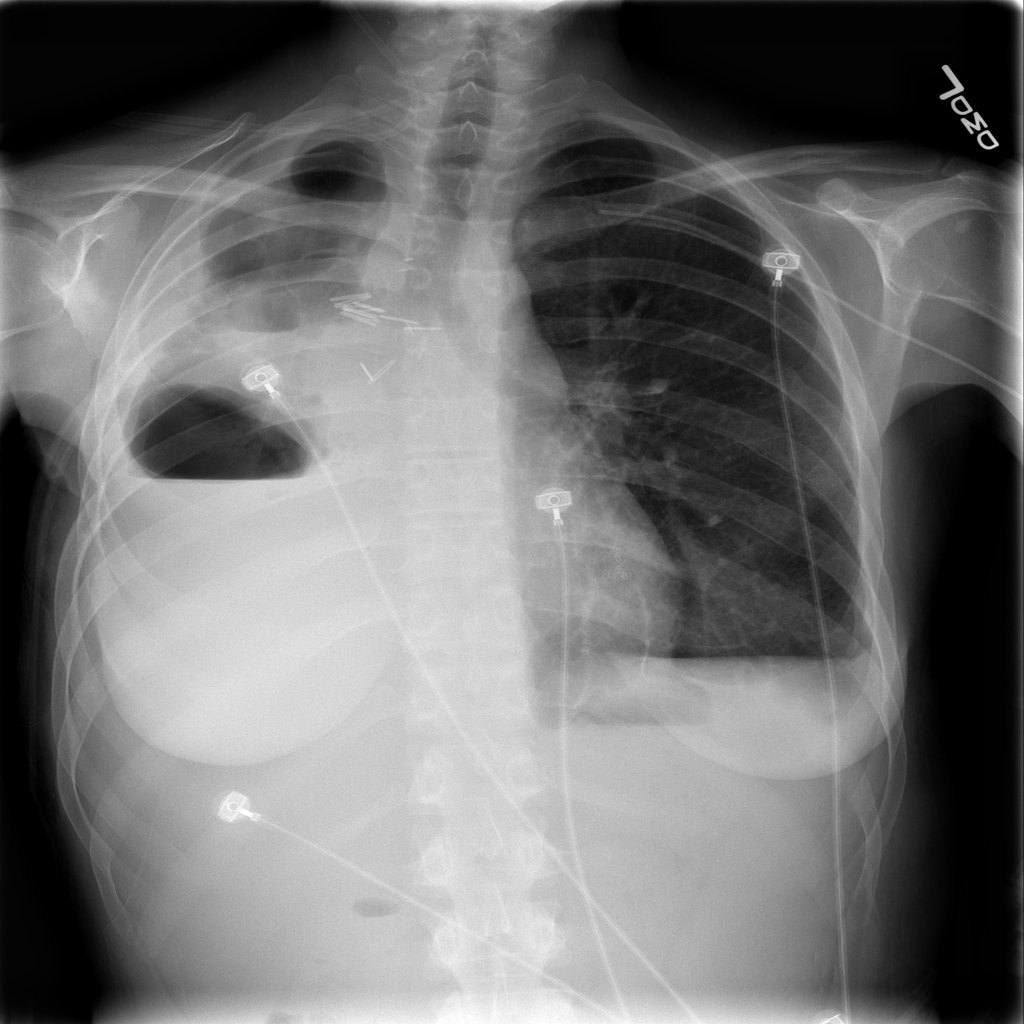
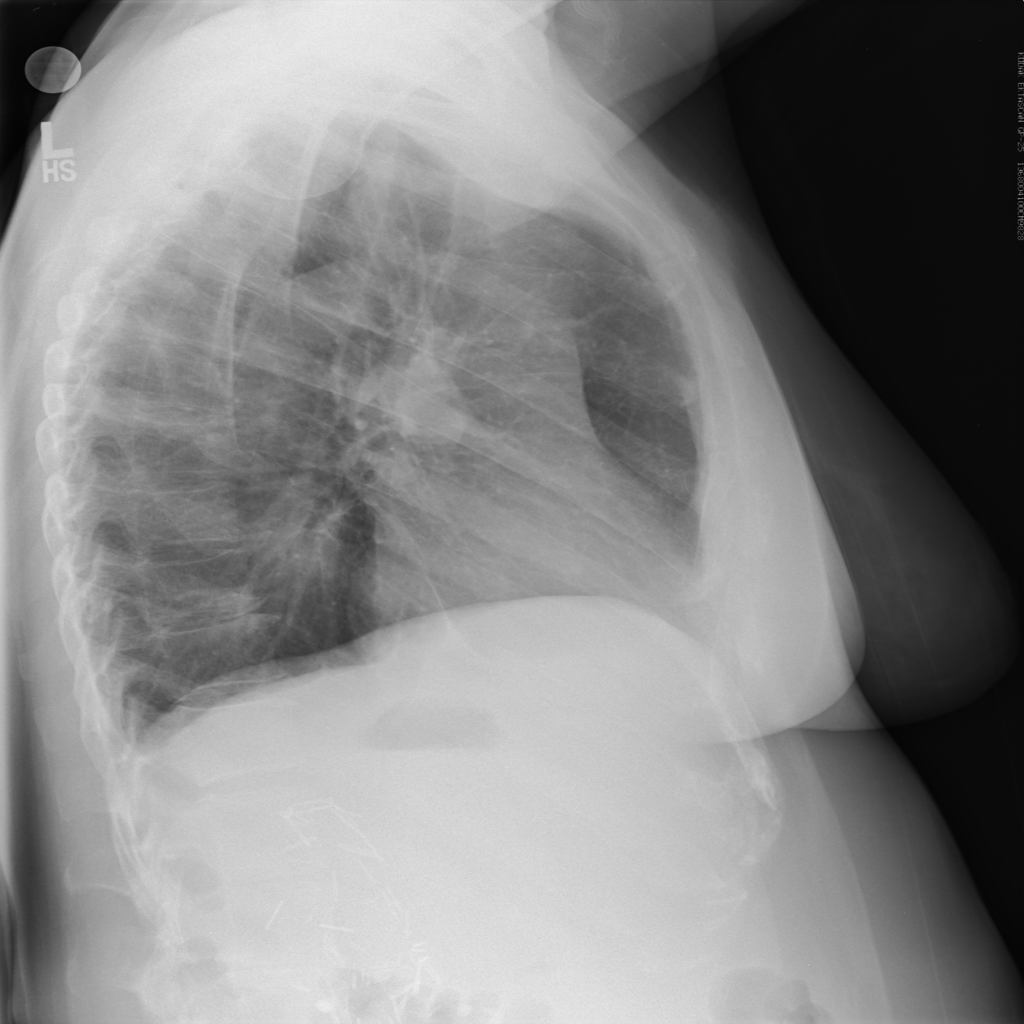
However, expert review of the dataset (ChestX-ray14) does not support this. In fact, there are significant concerns that the labeling of the dataset is a good deal weaker. I’ll just pick out two examples above that show a patient likely post R lobectomy with attendant findings classified as “No Findings” and the lateral chest X-ray which doesn’t even belong in the study database of all PA and AP films. These sorts of findings aren’t isolated – Dr. Luke Oakden-Rayner addresses this extensively in this post, from which his own observations are garnered below:
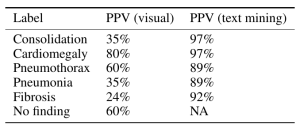
His final judgment is that the ChestX-ray14 dataset is not fit for training medical AI systems to do diagnostic work. He makes a compelling argument, but I think it is primarily a labelling problem, where the proposed 90% acccuracy on the NLP data mining techniques of Wang et al does not hold up. ChestX-ray14 is a useful dataset for the images alone, but the labels are suspect. I would call upon the NIH group to address this and learn from this experience. In that light, I am surprised that the system did not do a great deal better than the human radiologists involved in Dr. Ng’s group’s study, and I don’t really have a good explanation for it.
The evaluation of CheXNet by these individuals should be recognized:
Luke Oakden-Rayner: CheXNet an in-depth review
Paras Lakhani : Dear Mythical Editor: Radiologist-level Pneumonia Detection in CheXNet
Bailint Botz: A Few thoughts about ChexNet
Copyright © 2017