Author’s Note: This was a fun side-project for the American College of Radiology’s Residents and Fellows Section. Judy Gichoya and I co-wrote the article. The original article was posted by Judy to Medium and appeared on HackerNoon. It was really an enlightening gathering of experts in the field. There is a small, but hopefully growing number of radiologists who are also deep learning practitioners.
Written by Judy Gichoya & Stephen Borstelmann MD
In December 2017 , we (radiologists both in training, staff radiologists and AI practitioners) discussed our role as knowledge experts in world of AI, summarized here https://becominghuman.ai/radiologists-as-knowledge-experts-in-a-world-of-artificial-intelligence-summary-of-radiology-ec63a7002329. For the month of January, we addressed the performance of deep learning algorithms for disease diagnosis , specifically focusing on the paper by the stanford group — CheXNet: Radiologist-Level Pneumonia Detection on Chest X-Rays with Deep Learning. We continue to generate a large interest in the journal club , with 347 people registered , 150 of whom signed on January 24th 2018 to participate in the discussion.
The paper has had 3 revisions and is available here https://arxiv.org/abs/1711.05225 . Like many deep learning papers that claim super human performance , the paper was widely circulated in the news media, several blog posts , on reddit and twitter.
Please note that the findings of superhuman performance are increasingly being reported in medical AI papers. For example, this article denotes that “Medical AI May Be Better at Spotting Eye Disease Than Real Doctors”
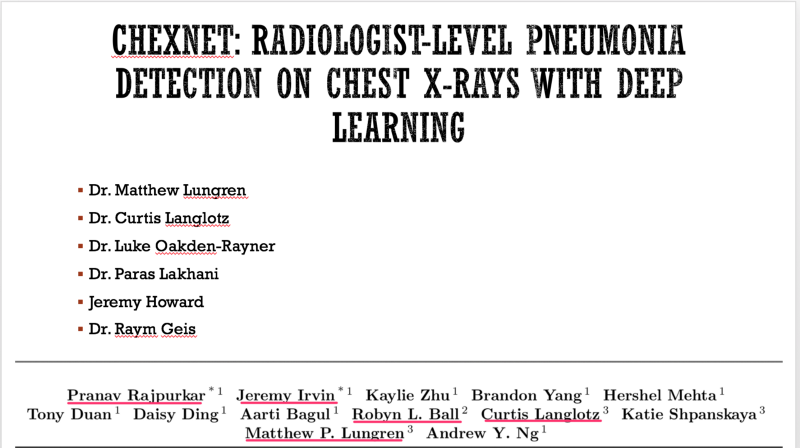
To help critique the ChexNet paper , we constituted a panel composed of the author team (most of the authors listed on the paper were kind enough to be in attendance — thank you!), Dr. Luke(blog) and Dr. Paras (blog) who had critiqued the data used and Jeremy Howard (past president and chief scientist of Kaggle, a data analytics competition site, Ex-CEO of Enlitic, a healthcare imaging company, and the Current CEO of Fast.ai, a deep learning educational site) to provide insight to deep learning methodology.
In this blog we summarise the methodology of reviewing medical AI papers.
Radiology 101
The ChexNet paper reviews performance of AI versus 4 trained radiologists in diagnosing pneumonia. Pneumonia is a clinical diagnosis — a patient will present with fever and cough , and can get a chest Xray(CXR) to identify complications of pneumonia. Patients will usually get blood cultures to supplement diagnosis. Pneumonia on a CXR is not easily distinguishable from other findings that fill the alevolar spaces — specifically pus , blood , fluid or collapsed lung called atelectasis. The radiologists interpreting these studies can therefore use terms like infiltrates , consolidation and atelectasis interchangeably.
Show me the data
The data used for this study is the ChestX-ray14 dataset which is the largest publicly available imaging data set that consists of 112,120 frontal chext xray radiographs of 30,805 unique patients and expands the ChestX-Ray 8, described by Wang, et. al. Each radiograph is labeled with one or more of 14 different pathology labels, or a ‘no finding’ label.
Labeling of the radiographs was performed using Natural Language Processing (NLP) by mining the text in the radiology reports. Individual case labels were not assigned by humans.
Critique: Labeling medical data remains a big challenge especially because the radiology report is a tool for communicating to ordering doctors and not a description of the images. For example , in an ICU film with a central line, tracheostomy tube and chest tube may be reported as “stable lines and tubes” without detailed description of the every individual finding on the CXR. This can be missclassified by NLP as a study without findings. This image-report disconcordance occurs at a high rate on this dataset.
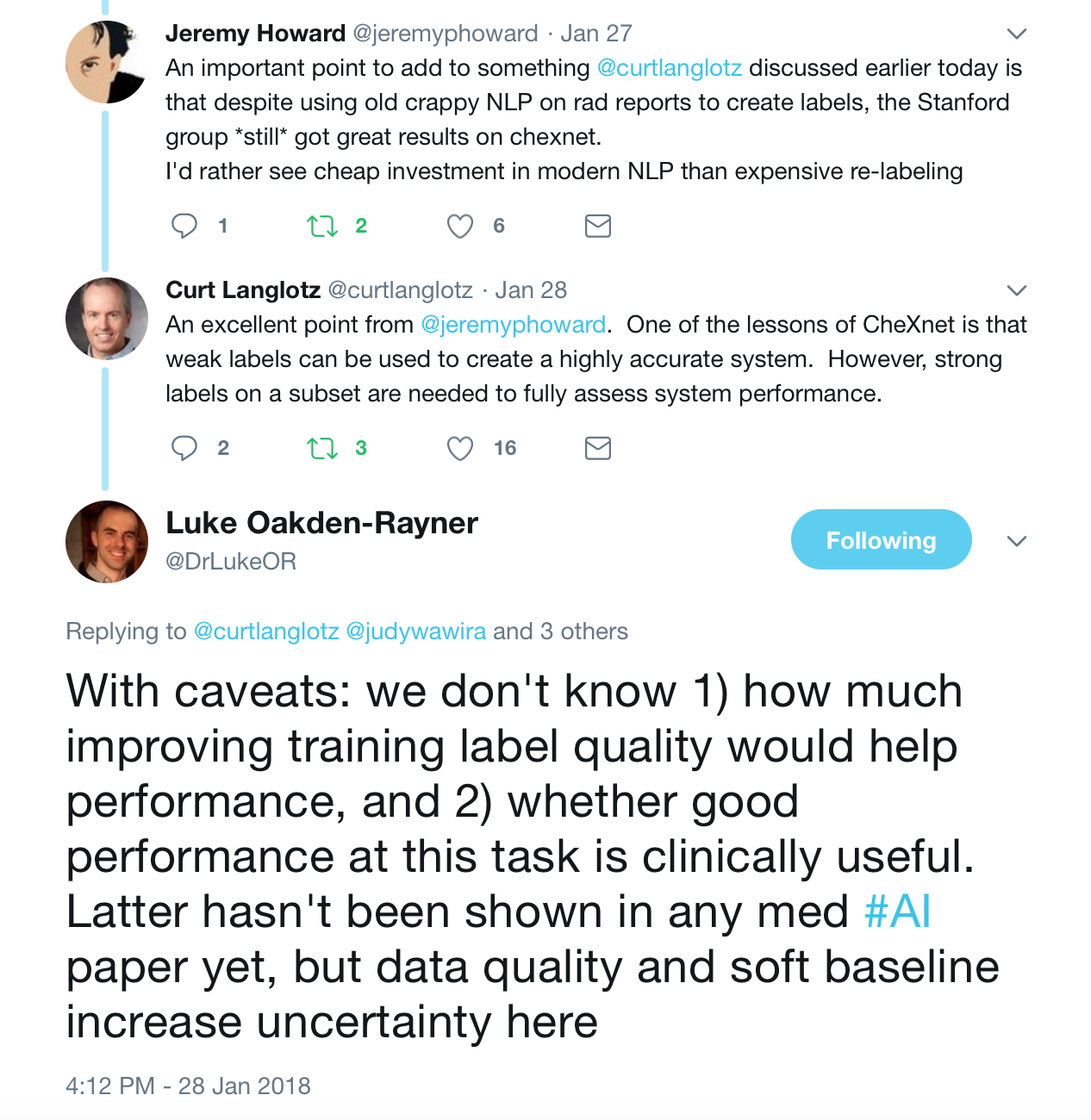
Moreover reportable findings could be ignored by the NLP technique and/or labeling schema, either through error or pathology outside of one of the 14 labels. The paper’s claims of 90%+ NLP mining accuracy do not appear to be accurate. (SMB,LOR,JH). One of the panelists — Luke reviewed several hundred examples and found the NLP labeling about 50% accurate overall compared to the image, with the pneumonia labeling worse — 30–40%.
Jeremy Howard notes that the use of an old NLP tool contributes to the inaccuracy due to the preponderance of ‘No Findings’ cases in the dataset skewing the data — he doesn’t think that the precision of normal findings in this dataset is likely improved over random. Looking at the pneumonia label, it is only 60% accurate. A lot of the discrepancy can be drawn back to the core NLP method, which he characterized as “massively out of date and known to be inaccurate”. He feels a re-characterization of the labels with a more up-to-date NLP system is appropriate.
The stanford group tackled the labeling challenge by having 4 radiologists (one specializing in thoracic imaging and 3 non thoracic radiologists) assign labels to a subset of the data for training created through a stratified random sampling, for a minimum of 50 positive cases of each label, with a final N=420.
Critique: The ChestXRay14 contains many patients with only one radiograph but those who had multiple studies tended to have many. While the text-mined reports may match clinical information, any mismatch between the assigned label and radiographic appearance hurts the predictive power of the dataset.
Moreover , what do the labels actually mean? Dr. Oakden-Rayner questions what the labels mean — do they mean a radiologic pneumonia or a clinical pneumonia? In an immunocompromised patient, radiography of a pneumonia might be negative, largely because the patient cannot mount an immune response to the pathogen. This does not mean that the clinical diagnosis of pneumonia is inaccurate. The imaging appearance and clinical appearance/diagnosis therefore would not match.
The closeness of four of the labels: Pneumonia, Consolidation, Infiltration, and Atelectasis introduces a new level of complexity. Pneumonia is a subset of consolidation and infiltration is a superset of consolidation. While the dataset labels these as 4 separate entities, to the radiologic practitioner they may not be separate at all. It is important to have experts look at images when doing an image classification task.
See a great summary of the data problems on this blog posting from Luke who was one of the panelists here.
Model
The CheXNet algorithm is a 121-layer deep 2D Convolutional Neural Network; a Densenet after Huang & Liu. The Densenet’s multiple residual connections reduce parameters and training time, allowing a deeper, more powerful model. The model accepts a vectorized two-dimensional image of size 224 pixels by 224 pixels.
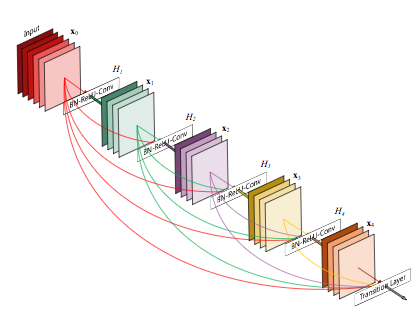
To improve trust in CheXNet’s output, a Class Activation Mapping (GRAD-CAM) heatmap was utilized after Zhou et al. This allows the human user to “see” what areas of the radiograph provide the strongest activation of the Densenet for the highest probability label.
Critique: Jeremy notes that image preprocessing of resizing to 224×224 pixel size images and adding random horizontal flips is fairly standard, but leaves room for potential improvement, as effective data augmentation is one of the best ways to improve a model. Image downsizing to 224×224 is a known issue — both from research and practical experience at Enlitic, larger images perform better in medical imaging (SMB: Multiple top 5 winners of the 2017 RSNA Bone age challenge had image sizes near 512×512). Mr. Howard feels there is no reason to leave Imagenet trained models this size any longer. Regarding the model choice, the Densenet model is adequate, but NasNets in the last 12 months have shown significant improvement (50%) over older models.
Pre-trained Imagenet weights were used, which is fine & a standard approach; but Jeremy felt it would be nice if we had a medical imagenet for some semi-supervised training of an AutoML encoder or a siamese network to cross validate patients — leaving room for improvement. Consider that Imagenet consists of color images of dogs, cats, planes and trains — and we are getting great results on X-rays? While better than nothing, ANY pretrained network trained on medical images in any modality would probably perform superiorly.
The Stanford team’s best idea was to train on multiple labels at the same time — it is best to build a single model that predicts multiple classes — counterintuitive, but bears out in deep learning models, and likely responsible for their model yielding better results than prior studies. The more classes you train the model on properly, the better results you can expect.
Results
F1 scores were used to evaluate both CheXNet model and the Stanford Radiologists.
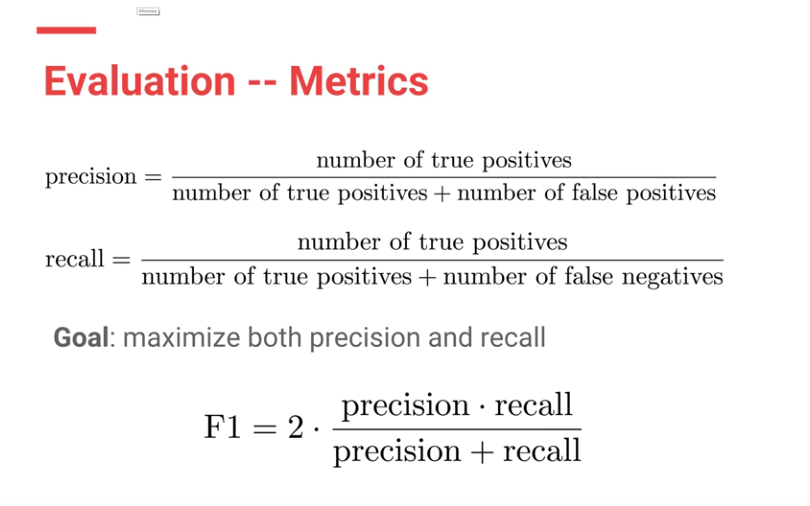
Each Radiologists’ F1 score was calculated by considering the other three radiologists as “ground truth.” ChexNet’s F1 score, was calculated vs. all 4 radiologists. A bootstrap calculation was added to yield 95% confidence intervals.
CheXnet’s results are as follows: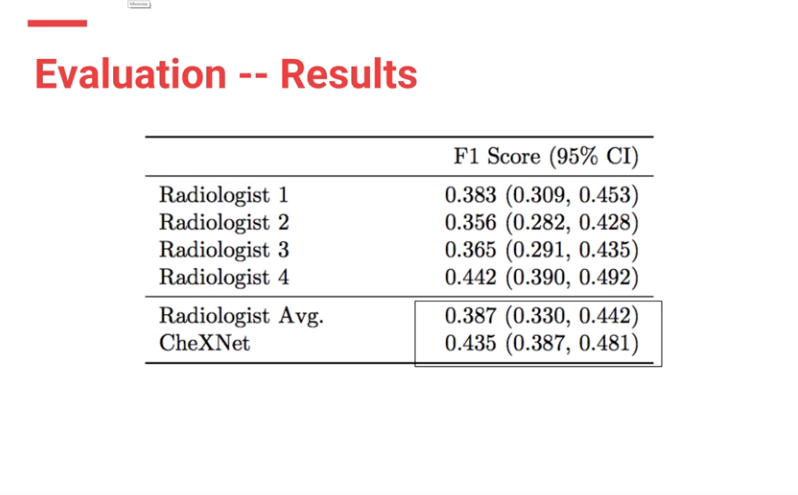
From the results, ChexNet outperforms human radiologists. The varying F1 scores can be interpreted to imply that for each study , 4 radiologists do not seem to agree with each other on findings. However there is an outlier (rad 4 — with an F score of 0.442) who is the thoracic trained radiologists who performs better than the ChexNet.
Moreover CheXNet has State of the Art (SOTA) performance on all 14 pathologies compared to prior publications.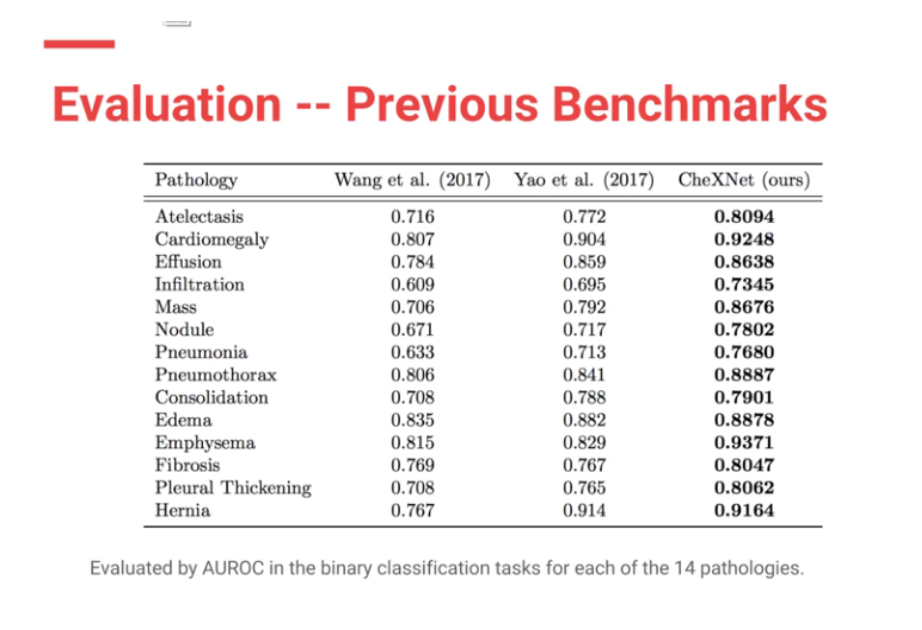
In my (JG) search , the Machine Intelligence Lab, Institute of Computer Science & Technology, Peking University, directed by Prof. Yadong Mu reports superior performance than the Stanford group. The code is open source and available here — https://github.com/arnoweng/CheXNet
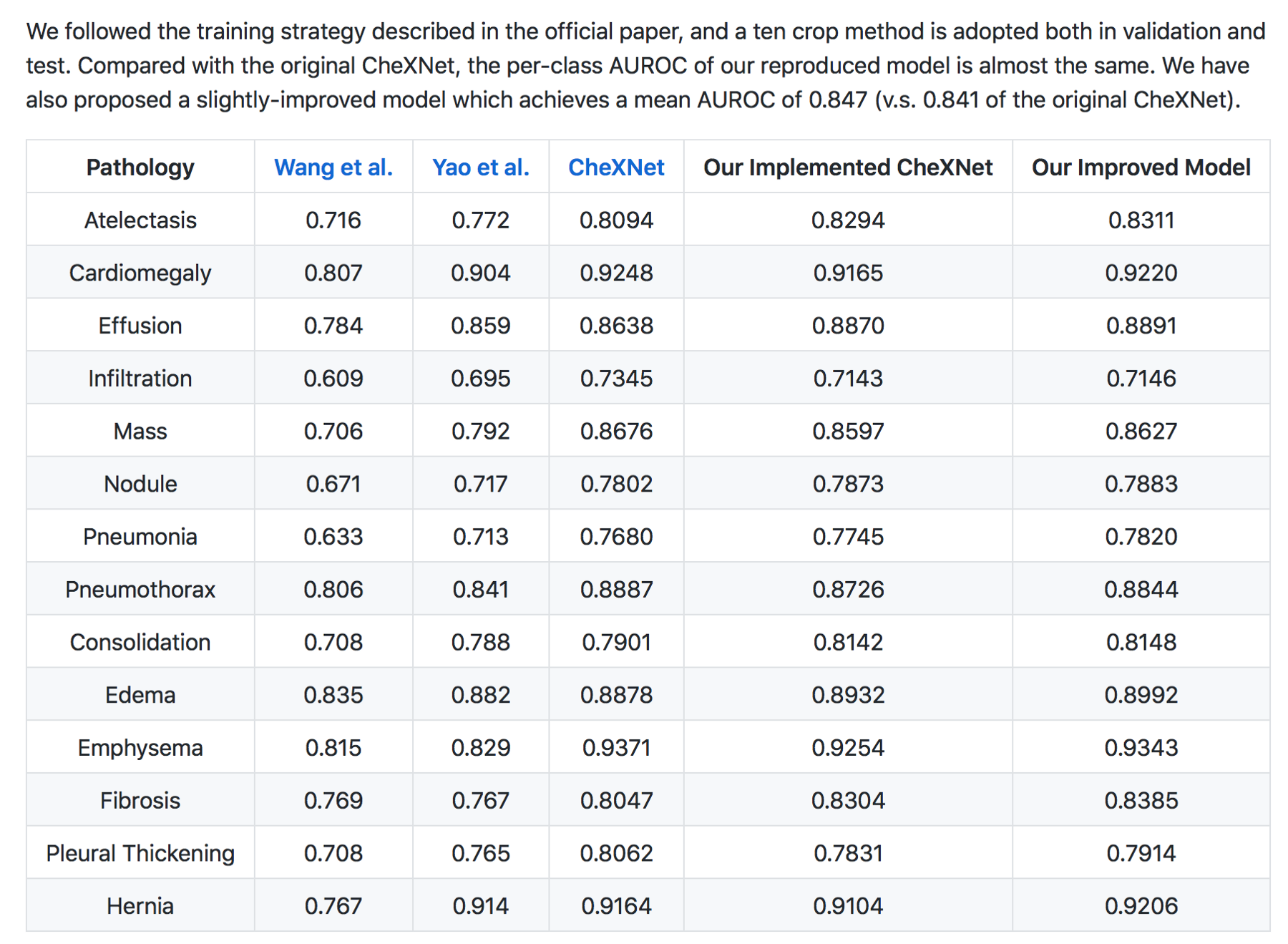
Critique — Various studies that assess cognitive fit show that human performance can be affected by lack of clinical information or prior comparisons that may affect their performance. Moreover, before the most recent version of the paper, human performance was unfairly scored against the machine.
Clinical significance
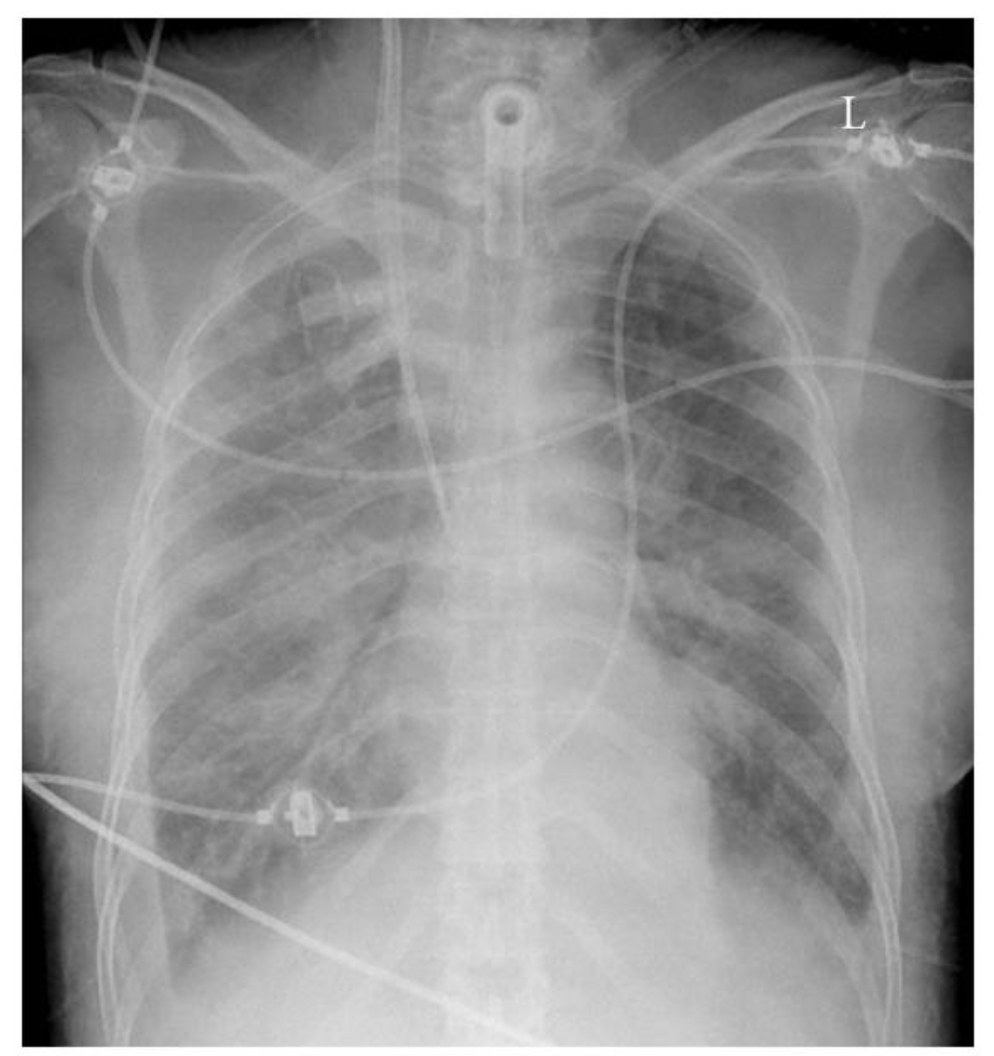
With the majority of labelled CXRs with pneumothorax having chest tubes present, the question must be raised: “are we training the Densenet to recognize pneumothoraces or chest tubes?”
Peer review
Luke Oakden-Rayner MD, a radiologist in Australia with expertise in AI & deep learning who was on our panel independently evaluated the ChestXRay-14 dataset, and CheXNet. He praises the Stanford team for their openness and patience in discussing the paper’s methodology, and their willingness to modify the paper to correct a methodologic flaw which biased against evaluating radiologists.
Summary
For the second AI journal club we analysed the pipeline of AI papers in medicine. You must make sure you are asking the right clinical question to be answered and not doing algorithms for the sake of doing something. Thereafter understand whether your data will help you answer the question you have, looking into details on how the data was collected and labeled.
To determine human level or super human performance, ensure the baseline metrics are adequate and not biased against one group.
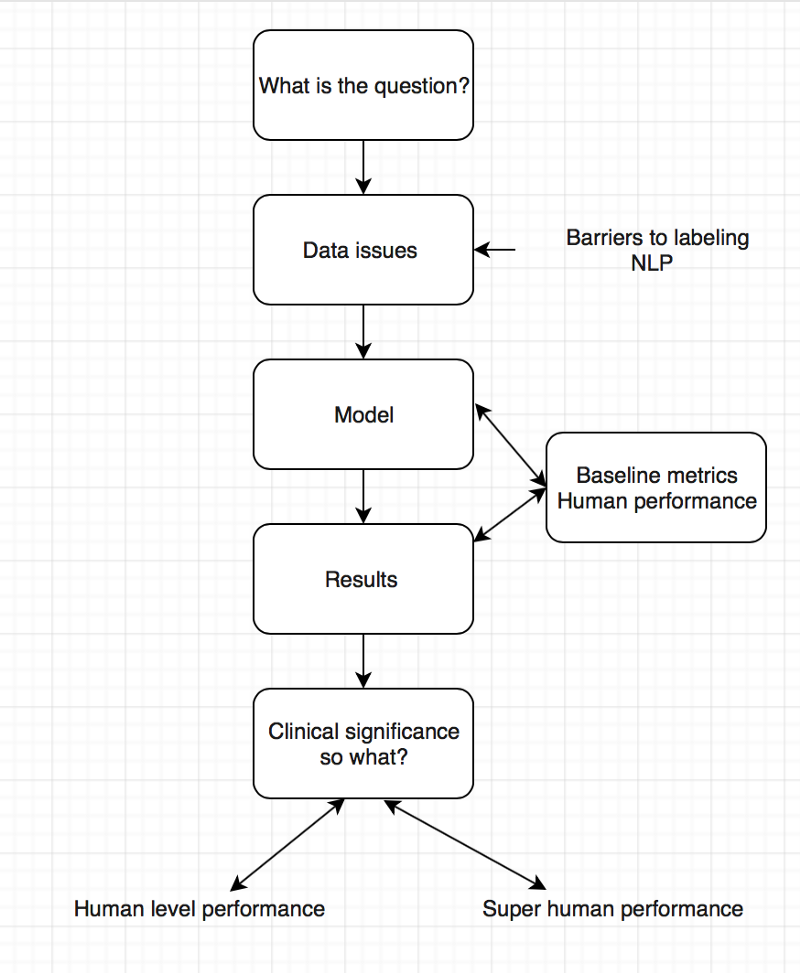
The model appears to give at-human performance for experts, or better than human performance for less-trained practitioners. This is in line with research findings and Enlitic’s experience. We should not be surprised by that; the research in Convolutional Neural Networks has consistently reported near-human or super-human performance consistently.
Take Aways
- There is exists a critical gap in the labeling of medical data.
- Do not forget the clinical significance of your results.
- Embrace peer review especially in medicine and AI
These were the best tweets regarding the problem of labeling medical data — aka do not get discouraged to attempt deep learning for medicine.
The journal club was a success, so if you are a doctor or an AI scientist , join us at https://tribe.radai.club to continue with the conversations on AI and medicine. You can listen to the recording of this journal club here : https://youtu.be/xoUpKjxbeC0 . Our next guest is Timnit Gebru who worked on US demographic household prediction using Google Street view images on 22nd February 2018. She will be talking on Using deep learning and Google Street View to estimate the demographic makeup of neighborhoods across the United States (http://www.pnas.org/content/114/50/13108).
Coming soon
For the journal club we developed a human versus AI competition for interepreting the CXRs in the dataset hosted at https://radai.club. We will be publishing the outcome of our crowdsourced labels soon, with a detailed analysis to check whether the model performance improves.
Say thanks
This I would like to thank the panelists including Jeremy Howard, Paras Lakhani, Luke Oakden-Rayner , and the Stanford ML team. Thanks to the ACR RFS AI advisory council members including Kevin Seals.
Article corrections made
- This article referred to Jeremy Howard (Ex-CEO of Kaggle) — updated to “president and chief scientist of Kaggle”
- Article stated NLP performance on that dataset is not likely improved over random.Jeremy clarified that the precision of the normal finding was what was not likely improved over random